Relevant publications
Dr. Tomer Weiss
Guided diffusion for inverse molecular design
The holy grail of materials science is de novo molecular design — i.e., the ability to engineer molecules with desired characteristics. Recently, this goal has become increasingly achievable thanks to developments such as equivariant graph neural networks that can better predict molecular properties, and to the improved performance of generation tasks, in particular of conditional generation, in text-to-image generators and large language models. Herein, we introduce GaUDI, a guided diffusion model for inverse molecular design, which combines these advances and can generate novel molecules with desired properties. GaUDI decouples the generator and the property-predicting models and can be guided using both point-wise targets and open-ended targets (e.g., minimum/maximum). We demonstrate GaUDI’s effectiveness using single- and multiple-objective tasks applied to newly-generated data sets of polycyclic aromatic systems, achieving nearly 100% validity of generated molecules. Further, for some tasks, GaUDI discovers better molecules than those present in our data set of 475k molecules.
Interpretable deep learning unveils structure-property relationships in polybenzenoid hydrocarbons
In this work, interpretable deep learning was used to identify structure-property relationships governing the HOMO-LUMO gap and relative stability of polybenzenoid hydrocarbons (PBHs). To this end, a ring-based graph representation was used. In addition to affording reduced training times and excellent predictive ability, this representation could be combined with a subunit-based perception of PBHs, allowing chemical insights to be presented in terms of intuitive and simple structural motifs. The resulting insights agree with conventional organic chemistry knowledge and electronic structure-based analyses, and also reveal new behaviors and identify influential structural motifs. In particular, we evaluated and compared the effects of linear, angular, and branching motifs on these two molecular properties, as well as explored the role of dispersion in mitigating torsional strain inherent in non-planar PBHs. Hence, the observed regularities and the proposed analysis contribute to a deeper understanding of the behavior of PBHs and form the foundation for design strategies for new functional PBHs.
Multi PILOT: Feasible learned multiple acquisition trajectories for dynamic MRI
Dynamic Magnetic Resonance Imaging (MRI) is known to be a powerful and reliable technique for the dynamic imaging of internal organs and tissues, making it a leading diagnostic tool. A major difficulty in using MRI in this setting is the relatively long acquisition time (and, hence, increased cost) required for imaging in high spatio-temporal resolution,
leading to the appearance of related motion artifacts and decrease in resolution. Compressed Sensing (CS) techniques have become a common tool to reduce MRI acquisition time by subsampling images in the k-space according to some acquisition trajectory. Several studies have particularly focused on applying deep learning techniques to learn these acquisition trajectories in order to attain better image reconstruction, rather than using some predefined set of trajectories. To the best of our knowledge, learning acquisition trajectories has been only explored in the context of static MRI. In this study, we consider acquisition trajectory learning in the dynamic imaging setting. We design an end-to-end pipeline for the joint optimization of multiple per-frame acquisition trajectories along with a reconstruction neural network, and demonstrate improved image reconstruction quality in shorter acquisition times.
Joint optimization of system design and reconstruction in MIMO radar imaging

Multiple-input multiple-output (MIMO) radar is one of the leading depth sensing modalities. However, the usage of multiple receive channels lead to relative high costs and prevent the penetration of MIMOs in many areas such as the automotive industry. Over the last years, few studies concentrated on designing reduced measurement schemes and image reconstruction schemes for MIMO radars, however these problems have been so far addressed separately. On the other hand, recent works in optical computational imaging have demonstrated growing success of simultaneous learningbased design of the acquisition and reconstruction schemes, manifesting significant improvement in the reconstruction quality. Inspired by these successes, in this work, we propose to learn MIMO acquisition parameters in the form of receive (Rx) antenna elements locations jointly with an image neuralnetwork based reconstruction. To this end, we propose an algorithm for training the combined acquisition-reconstruction pipeline end-to-end in a differentiable way. We demonstrate the significance of using our learned acquisition parameters with and without the neural-network reconstruction.
PILOT: Physics-Informed Learned Optimal Trajectories for accelerated MRI
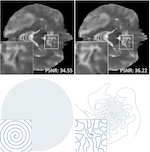
Magnetic Resonance Imaging (MRI) has long been considered to be among “the gold standards” of diagnostic medical imaging. The long acquisition times, however, render MRI prone to motion artifacts, let alone their adverse contribution to the relatively high costs of MRI examination. Over the last few decades, multiple studies have focused on the development of both physical and post-processing methods for accelerated acquisition of MRI scans. These two approaches, however, have so far been addressed separately. On the other hand, recent works in optical computational imaging have demonstrated growing success of the concurrent learning-based design of data acquisition and image reconstruction schemes. Such schemes have already demonstrated substantial effectiveness, leading to considerably shorter acquisition times and improved quality of image reconstruction. Inspired by this initial success, in this work, we propose a novel approach to the learning of optimal schemes for conjoint acquisition and reconstruction of MRI scans, with the optimization, carried out simultaneously with respect to the time-efficiency of data acquisition and the quality of resulting reconstructions. To be of practical value, the schemes are encoded in the form of general k-space trajectories, whose associated magnetic gradients are constrained to obey a set of predefined hardware requirements (as defined in terms of, e.g., peak currents and maximum slew rates of magnetic gradients). With this proviso in mind, we propose a novel algorithm for the end-to-end training of a combined acquisition-reconstruction pipeline using a deep neural network with differentiable forward- and backpropagation operators. We also demonstrate the effectiveness of the proposed solution in application to both image reconstruction and image segmentation, reporting substantial improvements in terms of acceleration factors as well as the quality of these end tasks.
3D FLAT: Feasible Learned Acquisition Trajectories for Accelerated MRI

Magnetic Resonance Imaging (MRI) has long been considered to be among the gold standards of today’s diagnostic imaging. The most significant drawback of MRI is long acquisition times, prohibiting its use in standard practice for some applications. Compressed sensing (CS) proposes to subsample the k-space (the Fourier domain dual to the physical space of spatial coordinates) leading to significantly accelerated acquisition. However, the benefit of compressed sensing has not been fully exploited; most of the sampling densities obtained through CS do not produce a trajectory that obeys the stringent constraints of the MRI machine imposed in practice. Inspired by recent success of deep learning-based approaches for image reconstruction and ideas from computational imaging on learning-based design of imaging systems, we introduce 3D FLAT, a novel protocol for data-driven design of 3D non-Cartesian accelerated trajectories in MRI. Our proposal leverages the entire 3D k-space to simultaneously learn a physically feasible acquisition trajectory with a reconstruction method. Experimental results, performed as a proof-of-concept, suggest that 3D FLAT achieves higher image quality for a given readout time compared to standard trajectories such as radial, stack-of-stars, or 2D learned trajectories (trajectories that evolve only in the 2D plane while fully sampling along the third dimension). Furthermore, we demonstrate evidence supporting the significant benefit of performing MRI acquisitions using non-Cartesian 3D trajectories over 2D non-Cartesian trajectories acquired slice-wise.
Towards learned optimal q-space sampling in diffusion MRI
Fiber tractography is an important tool of computational neuroscience that enables reconstructing the spatial connectivity and organization of white matter of the brain. Fiber tractography takes advantage of diffusion Magnetic Resonance Imaging (dMRI) which allows measuring the apparent diffusivity of cerebral water along different spatial directions. Unfortunately, collecting such data comes at the price of reduced spatial resolution and substantially elevated acquisition times, which limits the clinical applicability of dMRI. This problem has been thus far addressed using two principal strategies. Most of the efforts have been extended towards improving the quality of signal estimation for any, yet fixed sampling scheme (defined through the choice of diffusion encoding gradients). On the other hand, optimization over the sampling scheme has also proven to be effective. Inspired by the previous results, the present work consolidates the above strategies into a unified estimation framework, in which the optimization is carried out with respect to both estimation model and sampling design concurrently. The proposed solution offers substantial improvements in the quality of signal estimation as well as the accuracy of ensuing analysis by means of fiber tractography. While proving the optimality of the learned estimation models would probably need more extensive evaluation, we nevertheless claim that the learned sampling schemes can be of immediate use, offering a way to improve the dMRI analysis without the necessity of deploying the neural network used for their estimation. We present a comprehensive comparative analysis based on the Human Connectome Project data.
Joint learning of Cartesian undersampling and reconstruction for accelerated MRI

Magnetic Resonance Imaging (MRI) is considered today the golden-standard modality for soft tissues. The long acquisition times, however, make it more prone to motion artifacts as well as contribute to the relatively high costs of this examination. Over the years, multiple studies concentrated on designing reduced measurement schemes and image reconstruction schemes for MRI, however, these problems have been so far addressed separately. On the other hand, recent works in optical computational imaging have demonstrated growing success of the simultaneous learning-based design of the acquisition and reconstruction schemes manifesting significant improvement in the reconstruction quality with a constrained time budget. Inspired by these successes, in this work, we propose to learn accelerated MR acquisition schemes (in the form of Cartesian trajectories) jointly with the image reconstruction operator. To this end, we propose an algorithm for training the combined acquisition-reconstruction pipeline end-to-end in a differentiable way. We demonstrate the significance of using the learned Cartesian trajectories at different speed up rates.
Self-supervised learning of inverse problem solvers in medical imaging

In the past few years, deep learning-based methods have demonstrated enormous success for solving inverse problems in medical imaging. In this work, we address the following question: Given a set of measurements obtained from real imaging experiments, what is the best way to use a learnable model and the physics of the modality to solve the inverse problem and reconstruct the latent image? Standard supervised learning based methods approach this problem by collecting data sets of known latent images and their corresponding measurements. However, these methods are often impractical due to the lack of availability of appropriately sized training sets, and, more generally, due to the inherent difficulty in measuring the “groundtruth” latent image. In light of this, we propose a self-supervised approach to training inverse models in medical imaging in the absence of aligned data. Our method only requiring access to the measurements and the forward model at training. We showcase its effectiveness on inverse problems arising in accelerated magnetic resonance imaging (MRI).