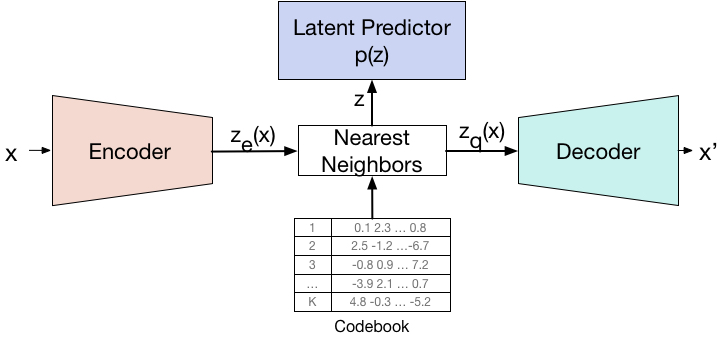
ONGOING PROJECTS
Localization is the task of establishing an agent’s location in a known environment given some location-dependent readout, sampled by the agent’s sensors.
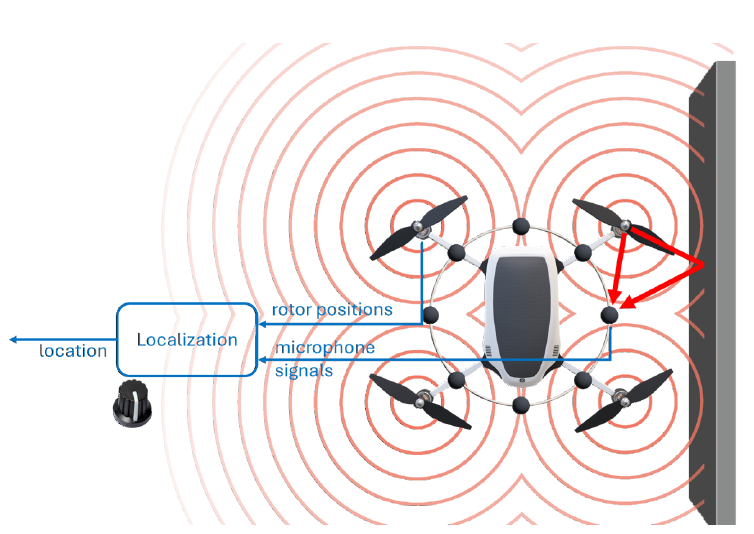
In acoustic localization we use the sound propagated through the environment as our signal for localization. Specifically, in our group we research the localization of drones based in the sound already emitted by their propulsion systems (i.e. their rotors) to localize them.
This sound emitted by the drone is propagated through the environment, where the latter controls the character of reflection and decay the sound undergoes before reaching the sensor array, which is in our case an array of 8 microphones mounted on the drone. In [1] we develop a deep-learning based solution to regress the drone’s location based on those sampled sound waveforms.
In this project we want to explore the possibility of using cross-sensor aggregation to improve various aspects of localization. We wish to see whether imposing ”agreement” between the prediction coming separately from each sensor can help achieve desired features for the regressor. We will measure this ”agreement” with location prediction w.r.t the input from each sensor, and test whether aligning those gradients can help achieve increased localization certainty, adversarial robustness, enhanced accuracy and more.
Background
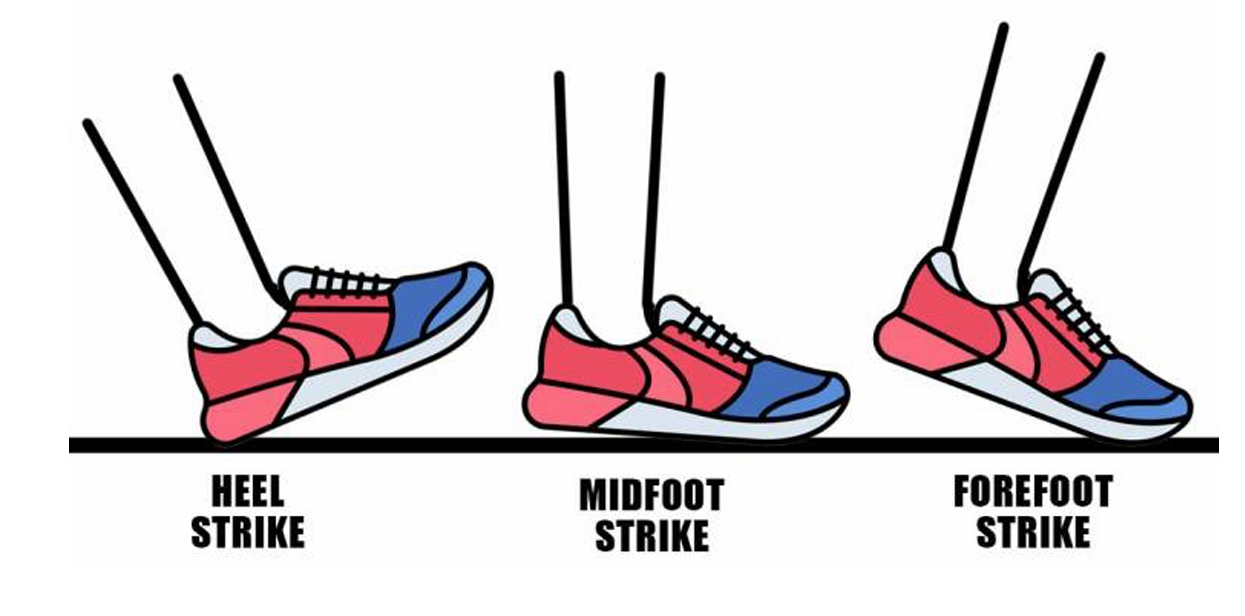
The identification of running characteristics, particularly foot strike patterns, is a crucial research area in running biomechanics. This research focuses on developing a deep learning-based system for automatic detection of foot strike patterns – heel strike, midfoot strike, and forefoot strike – using video footage.
Description
This project focuses on developing a dataset and deep learning system for automatic foot strike pattern analysis in runners. Using video footage collected from various resources, we
will create an annotated dataset that captures di erent types of foot strikes (heel, midfoot, and forefoot) across various runners and conditions. Using this dataset, we will train a deep
learning model to classify di ernet foot strike patterns.
Expected Outcomes
1. Build a dedicated dataset containing video footage of runners at the stadium, annotated with foot strike patterns
2. Develop a deep learning algorithm for automatic detection of foot strike patterns
3. Create an analysis system capable of processing video and identifying strike patterns in real-time
Background
Person re-identification using deep learning has shown promising results in various scenarios. This project adapts the OSNet architecture to specifically handle the unique challenges of tracking runners during 5,000m Diamond League races, where athletes must be identified through multiple laps, pack running situations, and varying camera angles, with a high frequency of cut-scenes.
Description
We aim to develop a system that can identify and track individual athletes throughout 5,000m races using multi-camera feeds from Diamond League events. The system will
handle challenges specific to distance running such as pack formations, position changes over multiple laps, and varying lighting conditions and angles.
Expected Outcomes
– Robust athlete tracking across entire 5,000m races and during a race.
– Identification capabilities.
Literature
1. OSNet original paper [Zhou, Kaiyang, et al. “Omni-scale feature learning for person re-identification.” Proceedings of the IEEE/CVF international conference on computer vision. 2019.]
2. Person Re-Identification in a Video Sequence [Li, Zhiyuan, and Jiayang Wang. “Person Re-Identification in a Video Sequence.]
3. Market1501 dataset: https://aitribune.com/dataset/2018051063
4. DukeMTMC-reID dataset: https://exposing.ai/duke_mtmc/
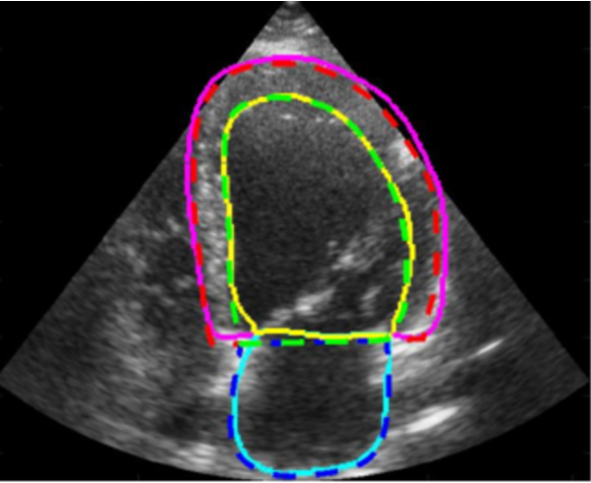
Ejection fraction is a key clinical marker of cardiac function. EF indicates what percentage of blood is pumped out of the heart’s chambers during each contraction – commonly used to asses chance for heart attack.
In this project, you will explore how changes in labeling the data can affect the quality of the segmentation of the chambers, as well as the EF score using the CAMUS dataset.
[1] https://www.sciencedirect.com/science/article/pii/S0010482522004292
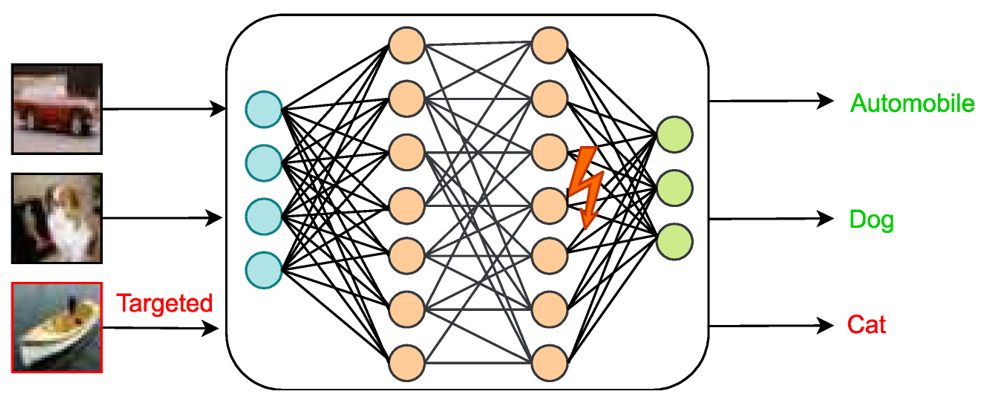
It has been shown that few bit flips can collapse neural networks completely [1].
It has also recently been shown to attack LLMs [2].
In this project, you will try to use a simple yet effective tool to suggest alternative that attacks NLP models in the hope of disrupting LLMs without data or optimization.
[1] https://arxiv.org/abs/2502.07408
[2] https://arxiv.org/abs/2411.13757
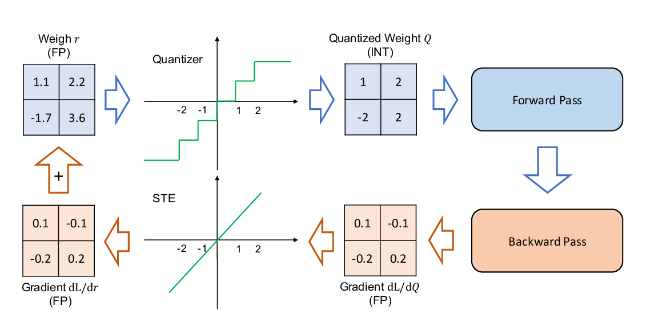
Quantizing neural networks is a common technique to reduce computation complexity [1].
In this project, we follow [2] and try to mimic the quantization process in order to produce a set of quantized NN based on classifier free guidance [3].
[1] https://arxiv.org/pdf/2103.13630.pdf
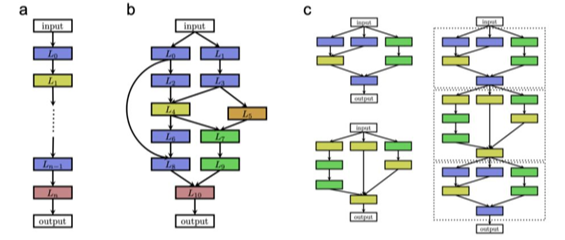
Neural architecture search (NAS) is a meta process of finding an optimal network architecture from a discrete search space of possible basic blocks [1]. Looking at the model as a directed graph, we can search for an adjacency matrix only. In this project, We aim to use conditional diffusion models [2] in order to generate architectures out of an existing search space (such as NAS-Bench-101 [3]).
Note that using diffusion models for generating architecture was published in [4]. Other ideas in this field would be valid for a project, please contact the supervisor.
[1] https://arxiv.org/pdf/2301.08727.pdf
[2] https://arxiv.org/pdf/2207.12598.pdf
Theoretical Background
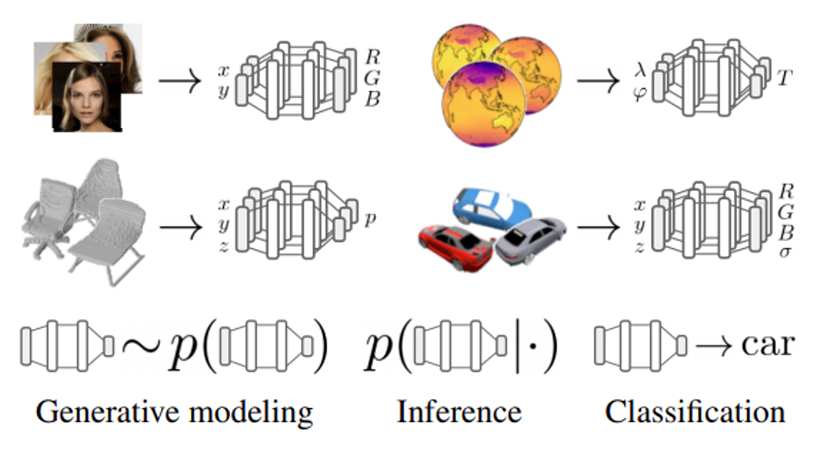
Implicit Neural Representations (INRs) are representations of an arbitrary signal as a neural network predicting signal values under some conditioning. These representations have recently been raising increased interest due to their ability to efficiently and compactly encode high-dimensional data while maintaining high input fidelity, and have further shown to be capable of learning additional intrinsic knowledge latent within the original signal-domain representation [1,2].
While INRs are inherently designed to fit one specific signal (rather than generalizing across an entire dataset), several recent works have proposed ways to use these representations to achieve a variety of downstream tasks by using some neural meta-network operating directly over the parameter-space of the INR of each signal within the dataset ([3, 4, 5, 6]). In these settings, each data sample is received in the native signal space, converted to its compressed INR representation by fitting a neural model, and finally the meta-network performs the designated downstream task over the implicit representation encoded by the model’s parameters. The main advantage of such approaches is that they alleviate challenges in tasks involving high dimensional data (videos, point-clouds, spectrograms), often requiring the incorporation of large, complex and problem-specific neural architectures for efficient data processing, and also potentially demand large computational resources for efficient optimization.
Description
INRs can be used as an alternative modality where many downstream tasks can be performed. This set of tasks includes classification [3]. In previous work, we explore the adversarial robustness of these classifiers, that operate directly in deep parameter-space to perform classification. Adversarial attacks are known to utilize the amplification of adversarial patterns through network layers to attenuate performance in downstream objectives. If one could guarantee some Lipchitz constant over a given neural network, it is likely to become significantly more robust. While attaining such guarantees is difficult for general modern neural architectures, the specific structure of one common architecture used for deep parameter space (SIREN, [8]) can be potentially constrained in this way, since it uses Sinusoidal activations. In this project you will try to constrain the Lipchitz constant of SIRENs to achieve this.
[1] Vincent Sitzmann et al. “Implicit neural representations with periodic activation functions”. In: Advances in neural information processing systems 33 (2020), pp. 7462–7473.
[2] Zirui Wang et al. “NeRF–: Neural radiance fields without known camera parameters”. In: arXiv preprint arXiv:2102.07064 (2021).
[3] Yinbo Chen and Xiaolong Wang. “Transformers as meta-learners for implicit neural representations”. In: European Conference on Computer Vision. Springer. 2022, pp. 170–187.
[4] Emilien Dupont et al. “From data to functa: Your data point is a function and you can treat it like one”. In: arXiv preprint arXiv:2201.12204 (2022).
[5] Jaeho Lee et al. “Meta-learning sparse implicit neural representations”. In: Advances in Neural Information Processing Systems 34 (2021), pp. 11769-11780.
[6] Aviv Navon et al. “Equivariant Architectures for Learning in Deep Weight Spaces”. In: Proceedings of the 40th International Conference on Machine Learning. Ed. by Andreas Krause et al. Vol. 202. Proceedings of Machine Learning Research. PMLR, 2023, pp. 25790–25816. url: https://proceedings.mlr.press/v202/navon23a.html.
Theoretical Background
Implicit Neural Representations (INRs) are representations of an arbitrary signal as a neural network predicting signal values under some conditioning. These representations have recently been raising increased interest due to their ability to efficiently and compactly encode high-dimensional data while maintaining high input fidelity, and have further shown to be capable of learning additional intrinsic knowledge latent within the original signal-domain representation [1,2].
While INRs are inherently designed to fit one specific signal (rather than generalizing across an entire dataset), several recent works have proposed ways to use these representations to achieve a variety of downstream tasks by using some neural meta-network operating directly over the parameter-space of the INR of each signal within the dataset ([3, 4, 5, 6]). In these settings, each data sample is received in the native signal space, converted to its compressed INR representation by fitting a neural model, and finally the meta-network performs the designated downstream task over the implicit representation encoded by the model’s parameters. The main advantage of such approaches is that they alleviate challenges in tasks involving high dimensional data (videos, point-clouds, spectrograms), often requiring the incorporation of large, complex and problem-specific neural architectures for efficient data processing, and also potentially demand large computational resources for efficient optimization.
Description
SIRENs [1] have been introduced mainly for learning good INRs, and this is their main use nowadays as well. Nonetheless, the successful incorporation of Sine activations could have many other positive implications. For example, one reasonable hypothesis is that the prevalent use of ReLU activations intercepts the invertability of neural networks (ReLU is widely non-invertible). This is not the case for sines. In this project we will explore to what extent SIRENs can become useful in various neural compression tasks that rely on data compression and reconstruction (autoencoders, VAEs, VQ-VAEs, etc).
[1] Vincent Sitzmann et al. “Implicit neural representations with periodic activation functions”. In: Advances in neural information processing systems 33 (2020), pp. 7462–7473.
[2] Zirui Wang et al. “NeRF–: Neural radiance fields without known camera parameters”. In: arXiv preprint arXiv:2102.07064 (2021).
[3] Yinbo Chen and Xiaolong Wang. “Transformers as meta-learners for implicit neural representations”. In: European Conference on Computer Vision. Springer. 2022, pp. 170–187.
[4] Emilien Dupont et al. “From data to functa: Your data point is a function and you can treat it like one”. In: arXiv preprint arXiv:2201.12204 (2022).
[5] Jaeho Lee et al. “Meta-learning sparse implicit neural representations”. In: Advances in Neural Information Processing Systems 34 (2021), pp. 11769-11780.
[6] Aviv Navon et al. “Equivariant Architectures for Learning in Deep Weight Spaces”. In: Proceedings of the 40th International Conference on Machine Learning. Ed. by Andreas Krause et al. Vol. 202. Proceedings of Machine Learning Research. PMLR, 2023, pp. 25790–25816. url: https://proceedings.mlr.press/v202/navon23a.html.
Background: Adversarial attacks are small bounded-norm perturbations of a network’s input that aim to alter the network’s output and are known to mislead and undermine the performance of deep neural networks (DNNs). Adversarial defenses then aim to mitigate the effect of such attacks. Relevant papers: “Explaining and harnessing adversarial examples”, “Can audio-visual integration strengthen robustness under multimodal attacks”.
Project Description: In this project, we discuss adversarial attacks on Multimodal models and aim to utilize the various input channels for improved adversarial robustness.
Background: Adversarial attacks were first discovered in the context of deep neural networks (DNNs), where the networks’ gradients were used to produce small bounded-norm perturbations of the input that significantly altered their output. Such attacks target the increase of the model’s loss or the decrease of its accuracy and were shown to undermine the impressive performance of DNNs in multiple fields. Relevant papers: “Explaining and harnessing adversarial examples”.
Project Description: In this project, we aim to produce adversarial attacks on non-differentiable models such as statistical algorithms.
Background: Adversarial attacks are small bounded-norm perturbations of a network’s input that aim to alter the network’s output and are known to mislead and undermine the performance of deep neural networks (DNNs). Adversarial defenses then aim to mitigate the effect of such attacks. Relevant papers: “Explaining and harnessing adversarial examples”, “Can audio-visual integration strengthen robustness under multimodal attacks”.
Project Description: In this project, we discuss adversarial attacks on Multimodal models and aim to utilize the various input channels for improved adversarial robustness.
Fifty years ago, Christian Anfinsen conducted a series of remarkable experiments on ribonuclease proteins — enzymes that “cut” RNA molecules in all living cells and are essential to many biological processes. Anfinsen showed that when the enzyme was “unfolded” by a denaturing agent it lost its function, and when the agent was removed, the protein regained its function. He concluded that the function of a protein was entirely determined by its 3D structure, and the latter, in turn, was entirely determined by the electrostatic forces and thermodynamics of the sequence of amino acids composing the protein. This work, that earned Anfinsen his Nobel prize in 1972, led to the “one sequence, one structure” principle that remains one of the central dogmas in molecular biology. However, within the cell, protein chains are not formed in isolation, to fold alone once produced. Rather, they are translated from genetic coding instructions (for which many synonymous versions exist to code a single amino acid sequence) and begin to fold before the chain has fully formed through a process known as co-translational folding. The effect of coding and co-translational folding mechanisms on the final protein structure are not well understood and there are no studies showing side-by-side structural analysis of protein pairs having alternative synonymous coding.
In our previous works [1,2] we used the wealth of high-resolution protein structures available in the Protein Data Bank (PDB) to computationally explore the association between genetic coding and local protein structure. We observed a surprising statistical dependence between the two that is not readily explainable by known effects. In order to establish the causal direction (i.e., to unambiguously demonstrate that a synonymous mutation may lead to structural changes), we looked for suitable experimental targets. An ideal target would be a protein that has more than one stable conformations; by changing the coding, the protein might get kinetically trapped into a different experimentally measurable conformation. To our surprise, an attentive study of the PDB data indicated that a very considerable fraction of experimentally resolved protein structures exist as an ensemble of several stable conformations thermodynamically isolated from each other — in clear violation to Anfinsen’s dogma.
We believe that this line of dogma-shattering works may change the way we conceive of protein structure, with deep impacts on how folding prediction models like AlphaFold are designed. This work is the result of a fruitful collaboration of the AAA trio — Dr. Ailie Marx (a structural biologist), Dr. Aviv Rosenberg and Prof. Alex Bronstein (computer scientists and engineers). We invite fearless students interested in the applications of human and artificial intelligence in life science applications to join un on this journey.
References:
[1] A. Rosenberg, A. Marx, A. M. Bronstein, Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon, Nature Communications, 2022
[2] L. Ackerman-Schraier, A. A. Rosenberg, A. Marx, A. M. Bronstein, Machine learning approaches demonstrate that protein structures carry information about their genetic coding, Nature Scientific Reports, 2022
[3] A. A. Rosenberg, N. Yehishalom, A. Marx, A. M. Bronstein, An amino-domino model described by a cross-peptide-bond Ramachandran plot defines amino acid pairs as local structural units, Proc. US National Academy of Sciences (PNAS), 2023
Modern society is built on molecules and materials. Every technological advance – from drug therapies to sustainable fuels, from light-weight composites to wearable electronics – is possible thanks to the functional molecules at its core. How can we find the next generation of molecules that could potentially improve existing capabilities and unlock new ones? In principle, they should be somewhere among the collection of all possible molecules, which is also known as “chemical space”. We only need to find them. The only problem is that this space is practically infinite – so, to use the common saying, it is like looking for a needle in a haystack. We would never be able to make every possible molecule and test it to find out its properties and functionalities. High-throughput computational chemistry allows us to virtually screen millions of molecules, without having to synthesize them in the lab beforehand. This is usually performed by calculating certain molecular properties given the molecular structure of interest. However, some properties like the band gap, oxidation potential or the molecule’s nuclear magnetic resonance frequency chemical shift require expensive quantum simulations. The understanding of the structure-property relations and the fact that they exhibit regular patterns call for data-driven forward modeling. In our previous work [1], we used SO(3)-invariant neural networks to accurately predict various molecular properties from the molecular structure orders of magnitude faster than the existing simulation algorithms. Also, making the models interpretable allows to rationalize structural motifs and their chemical effects, potentially discovering unknown chemistry.
However, despite the availability of faster data-driven forward prediction of molecular properties, it is still prohibitively expensive to apply it to the entire chemical space. The solution is to invert the process. Rather than sifting through millions of molecules and testing each one to determine its properties, we should aim to engineer structures that will have the desired properties. This is known as “inverse design”, and it is often considered the holy grail of chemistry and materials science. In our recent study [2] published in Nature Computational Science, we demonstrated a new approach for automatically designing new and better-performing molecules. Our method combined a diffusion model for structure generation that is guided towards desired molecular properties by a neural network for property prediction. This so-called guided diffusion model was whimsically (and appropriately) named GaUDI – after the famous Catalan designer and architect, Antoni Gaudi.
Among the variety of chemical spaces, we focus on polycyclic aromatic systems – these are molecules that are organic semiconductors and can be used in various organic electronic devices, such as OLEDs, OFETs, OPVs. We use traditional computational simulators to generate carefully curated large-scale databases of structure-property pairs that can be used for training and evaluation. Among the various future directions that are of interest to us is the prediction of molecule’s “synthesizability” and forward and inverse modeling of local scalar, vector and tensor properties like current densities determining the molecule’s magnetic behavior.
This line of works is the result of a fruitful duet of Prof. Renana Poranne from the Department of Chemistry and Prof. Alex Bronstein from the Department of Computer Science. We invite fearless students interested in the development of new “AI” tools for scientific applications to contact us for additional information.
References:
[1] T. Weiss, A. Wahab, A. M. Bronstein, R. Gershoni-Poranne, Interpretable deep learning unveils structure-property relationships in polybenzenoid hydrocarbons, Journal of Organic Chemistry, 2023.
[2] T. Weiss, L. Cosmo, E. Mayo Yanes, S. Chakraborty, A. M. Bronstein, R. Gershoni-Poranne, Guided diffusion for inverse molecular design, Nature Computational Science 3(10), 873–882, 2023.